Genome sequencing

What is Genome Sequencing?
Genome sequencing or DNA sequencing refers to the process of determining the complete sequence of nucleotides in genome (read DNA). Ignoring the 3 dimensional orientation of the DNA strand, DNA can be thought of as a linear sequence of 4 nucleotide base pairs (bp), namely Adenine (A), Thymine (T), Guanine (G), Cytosine (C). A pairs with T and G pairs with C. Sequencing allows us to determine the sequence such as AATGCCAT….
Why is sequencing important?
Have you heard of Precision Medicine (PM)?
Think about it this way, everytime you face a serious medical issue, instead of prescribing generic medicine/treatment procedures which is given to all the patient with similar symptoms, you get a prescription tailored for you, by analysing your genetic code and symptoms. The exact same treatment for exact same symptoms on me and you might have different outcomes, it may not be pleasant for one of us.
Now to enable precision medicine, one of the key steps is to create profile for the patient, which is where sequencing comes into picture. Therfore it is important to invest and develope sequencing methods which have higher accuracies and are faster and cheaper.
check out this video for one success story of precision medicine
How is sequencing done?
Attempts to sequence DNA dates back to 1970s (wikipedia link to brief history). Intital techniques, even though impressive were quite costly and terribly slow for real world applications such as using it for human genome. Humans have come a long way now, both time and costs to sequence whole genome has come down drastically and now it’s practical to think about genome sequencing as part of diagnostics.
Recent techniques are known as NGS (Next Generation Sequencing) or HTS (High Throughput Sequencing). In the following I will discuss one successful commercial technique used today. While reading up on this topic I realised that the literature around this topic is presented with alot of fancy words (that made no sense to me and will probably not make sense to average joe). But then the topic is quite specific and not meant for avaerage joe, however to grasp things I found it helpful to look around for the meaning of fancy words as they help in providing context and fill in the gaps. I will try to present it in a simple language with links to resources where necessary.
DNA Nanoball Sequencing
Many of the sequencing method will broadly involve steps such as
- splitting the DNA strand into smaller pieces
- generating multiple copies of the smaller pieces
- processing the cluster of smaller pieces with marker nucleotides which are fluorescent and are color coded depending upon the nucleotide
- capturing the image/data
- bioinformatics on data to extract sequences
The specifics of each of the steps depend upon the method used. In DNA Nanoball Sequencing the longer DNA strands are sonicated (using sound) to break them into smaller pieces, then pieces of appropriate lengths (typically 400-500 bp) are chosen for futher processing (how it’s done is irrelevant for the conceptual big picture).
Here is a cool video showing the sonification process
Nest step involves attaching adapter sequences and to convert the small fragments into curcular struture which is then replicated by a procedure call Rolling circle replication. The output of this step leads to lot of circular copies which when linearised, forms a long strand of DNA, because of the adapter sequences (desinged to be “sticky”) the long strand folds onto itself making a ball like structure which is about 300nm in diameter. These nanoballs are then attached to a specially constructed microarray, nanoballs are arranged in highly ordered fashion hence are densly packed which is what allows the massive parallel processing. One way to think about these arrays:
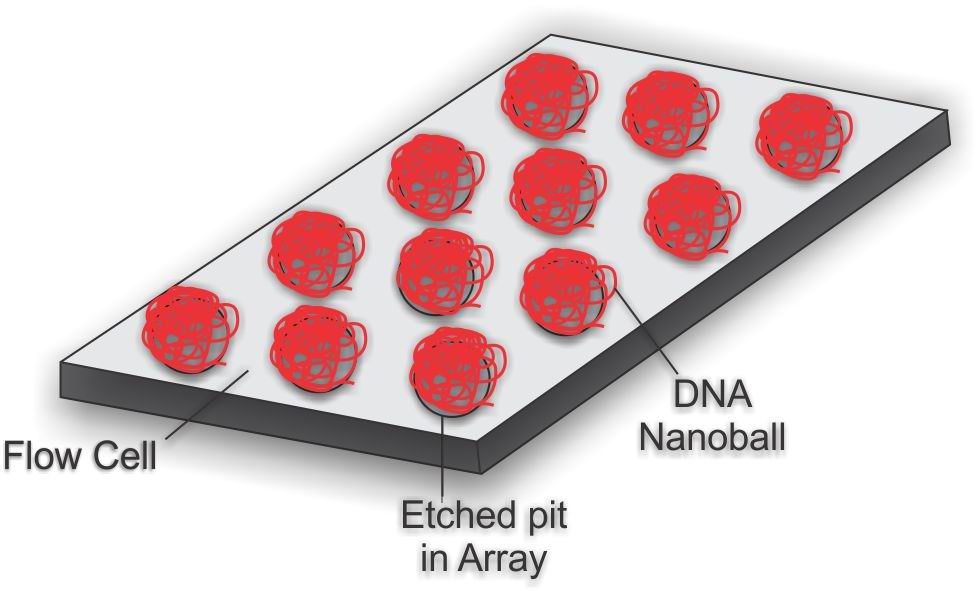
The sequencing part is easier to analyse with the unchained visualisation, that is consider the image of long DNA strand from the nanoball, we know the adapter sequences which we attached earlier, so we can construct an “anchor” sequence which will attach to the adapter part of the long template strand.
Now we introduce, something called “probes”, these are short sequences with “degenerate (not important)” nucleotides in all but one position, and have a color coded floroscence depending upon that one non-denerate nucleotide (for example in the image, A is red, C is yellow, G is green, T is blue).
Anchor is attached to the adapter sequence, the microarray cell is flooded with one type of probes (say non-degenrate nucleotide at one position to the left of adapter part), only the probe containing the complimentary non-degenrate nucleotide will bind, we can wash away the redundant chemicals and record the florocent signals, the color will indicate which one of the A/T/G/C was present at that location. We can wash away the anchor and probe of this round and repeat the process with probes of different locations. As described nicely in this image and its caption :
 1. Single strand of DNA from a DNA nanoball 2. Attach complementary anchor to known adapter sequence 3. Add probe set #1 and DNA Ligase to interrogate position #1 to the left of the anchor. Only the complementary probe binds 4. Wash away all unbound probes and Ligase, and read the color of the fluorophore 5. Denature anchor and probe from DNA nanoball and add another anchor 6. Add probe set #2 and DNA ligase to interrogate position #2 to the left of the anchor. Repeat.
1. Single strand of DNA from a DNA nanoball 2. Attach complementary anchor to known adapter sequence 3. Add probe set #1 and DNA Ligase to interrogate position #1 to the left of the anchor. Only the complementary probe binds 4. Wash away all unbound probes and Ligase, and read the color of the fluorophore 5. Denature anchor and probe from DNA nanoball and add another anchor 6. Add probe set #2 and DNA ligase to interrogate position #2 to the left of the anchor. Repeat.
This is an iterative process, where after every round of probe/ligation step, excessive chemicals are washed and the fluorocence is captured in form of a image. These images are then analysed to infer the sequence of the template strand using the colour coding scheme of the nucleotides.
I chose this method because it is at the crux of commecial product by Beijing Genomics Institute (BGI). Their method is known as Combinatorial probe anchor synthesis (cPAS), the technology is capable of processing around 40 million nucleotides per second (as of 2018, which is a remarkable number)
References
- http://www.genomenewsnetwork.org/resources/whats_a_genome/Chp2_1.shtml
- https://www.ebi.ac.uk/training/online/course/ebi-next-generation-sequencing-practical-course/what-you-will-learn/what-next-generation-dna-
- https://en.wikipedia.org/wiki/DNA_sequencing
- https://en.wikipedia.org/wiki/DNA_nanoball_sequencing
- http://www.seq500.com/en/portal/Seq-500.shtml
- https://en.wikipedia.org/wiki/Sonication
- https://www.creative-biogene.com/blog/index.php/2016/11/01/brief-introduction-on-three-generations-of-genome-sequencing-technology/
- http://koreabizwire.com/scientists-find-whole-genome-sequencing-of-korean-individual/67503
